We study the evolution and genetics of plant-microbe symbiosis!

Soybean field and farmstead near the KBS LTER; Photo Credit: GP Robertson, Michigan State University
Check out GEMS, our Biological Integration Institute on Genomics and Eco-Evolution of Multiscale Symbiosis!

We do a lot of work on mutualism evolution. Why study mutualism evolution?

Mesocosms of grasses and clover in our greenhouse
Mutualisms (mutually-beneficial species interactions) are everywhere! The swapping of resources between species is a fundamental force in biology. Your gut microbes help you digest your food, and in return they get to eat some of it and have a cozy place to hang out. The reason soybeans and beans are good for you is because these plants form special associations with nitrogen-fixing rhizobia, which provide their host plants with lots of nitrogen to enrich their seeds with protein.

Soybeans in our greenhouse
Despite the importance of mutualisms to natural ecosystems, agriculture, and human health, their evolution is not well-understood. How do mutualisms arise in the first place? What evolutionary forces are important in ongoing mutualism coevolution? Over deeper timescales, what generates phylogenetic diversity in mutualisms? These are very big and unresolved questions in mutualism evolution. Currently my group is using a wide variety of methods to address questions about contemporary coevolution in mutualisms using legume-rhizobium interactions as our study system. We tend to use genetic models because they are tractable in the lab and we can effectively merge ecologically-relevant (fitness-related) traits with genomic data to understand the mechanistic basis of coevolution.
① Genotype X Environment Interactions
We are interested in how the environment shapes the mutualism between legumes and rhizobia. There is a strong connection between the mutualism we study here and the environmental factors which exist in the world. We have found through several studies that factors such as nitrogen addition, drought, light availability, and herbivory all impact the mutualism. Our current projects work to understand the impact of artificial selection on mutualism and the impact of partner quality degradation on communities.
Amanda works on understanding the role that domestication of soybeans has on the mutualism with the microbial community. Her work looks into the role that human influence has played in selection of traits and how those may have impacted the symbiosis.
Natalie works to understand how ecosystem-level processes such as nitrogen cycling in soils respond to long term nitrogen fertilization and drought/flooding, and how these complex effects influence plant-microbe interactions.
② Genotype X Genotype Interactions
Genotype-by-genotype interactions occur when the phenotypic effects of one partner’s genotype depend on the genotype in the other interacting partner. We are interested in incorporating this line of thought into how biological nitrogen fixation, and other symbiotic benefits that rhizobium provide, can be impacted by soil nitrogen in long term studies. We are interested in learning more about the relative fitness of both host and partner as they interact in changing environments.
Jason studies the co-evolution of symbiosis using the legume-rhizobia model, specifically investigating how additional nitrogen treatment influences the plant host and their associated symbionts. His main question is trying to understand how different nitrogen levels affect the symbiotic traits across multiple hosts.
Megan wants to identify how the microbial interactions of sinorhizobium in non-nodulating hosts such as avena (oat) influence medicago plant health. Her work looks at the interactions between these plant species and symbiont and how both species can impact the evolution of the mutualism and selection on sinorhizobium when in symbiosis.
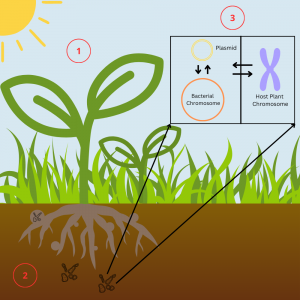
③ Mobile Genetic Elements
Mobile genetic elements are segments of genetic material which are able to move within or between genomes. MGEs are of interest because of the role they play in the symbiosis. We have found evidence of partner quality variation and the genes responsible for symbiosis on plasmids. We are currently interested in plasmid biology including mutation, structure, evolution, and phylogeny.
David studies how symbiosis plasmid mutations at particular genes and/or entire plasmids can alter partner quality in rhizobium in response to high nitrogen environments.
Ivan aims to understand how genetic variation in symbiotic microbes affects, maintains and allows symbiotic relationships in leguminous plants and their rhizobial and other endophytic partners.
Sierra studies plasmid-chromosome coevolution in a population of rhizobium undergoing nitrogen deposition. She wants to be able to determine whether or not the cost of carrying plasmids is affected by environmental conditions.