
Research Interests
Much genetic diversity and evolutionary innovation are in the tropics, yet tropical plants remain under-explored in their basic biology and their enormous potential for novel discovery. Our group has been focusing on genome evolution and reproductive biology of selected tropical crop plants. The goal of our research program is to understand the genetic and genomic basis of reproduction and biomass yield of these target species. Our current research interests include understanding the evolution of sex chromosomes in the family Caricaceae and the molecular basis of sex determination in papaya, exploring the genomic basis of biomass yield in sugarcane, and unraveling the genome structure and the dynamics of genome evolution in papaya, sugarcane, and coffee.
Evolution of sex chromosomes in Caricaceae
Papaya is among the limited number of plant species that are trioecious with three sex forms – female, male, and hermaphrodite. In the two breeding systems of papaya, it is either dioecious with male and female or gynodioecious with hermaphrodite and female. Sex determination in papaya is controlled by nascent sex chromosomes with a small male specific region at 8.1 Mb, about 10 – 15% of the Y chromosome. Several dioecious and trioecious species of Vasconcellea, a related genus of Carica, were studied, showing that sex chromosomes have evolved in those species as well. Papaya and its relatives in the Caricaceae family offer a rare opportunity to use genomics for studying the evolution of sex chromosomes, and the evolution of genes that reside on them.
Sex determination in papaya is likely controlled by two genes on the MSY, one promoting stamen development and the other suppressing carpels, as supported by the co-existence of gynodioecious (female and hermaphrodite) and dioecious (male and female) papaya. Papaya male flowers have 10 stamens and a trace of an aborted pistil. Female flowers have a large functional pistil, but are entirely devoid of stamens. The abortion of stamens in female flowers occurs at inception, whereas the abortion of carpels in male flowers occurs at a much later developmental stage. We have identified candidate genes for sex determination, and experiments are underway to transform these genes into target sex types for function complementation.
The extent of sex chromosome evolution in other dioecious and trioecious species of Caricaceae is being investigated.
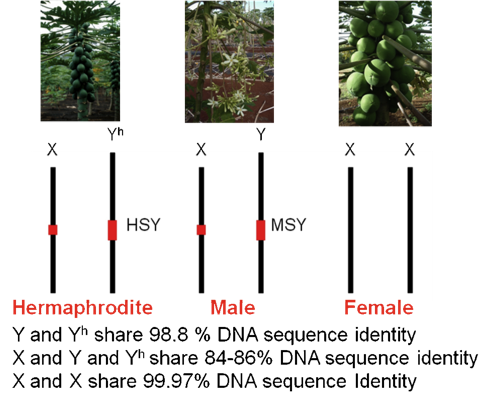
Figure 1. The genotypes of sex chromosomes in three sex types of papaya.
Genome evolution in Caricaceae
Papaya has a number of characteristics that contribute to its being used as an experimental model for tree crops. Papaya has small genome of 372 Mb, a short juvenile phase of 3 to 8 months, and a short generation time of 9 to 15 months. Flowering and fruiting are continuous throughout the year with the production of one to three ripe fruit per week and hundreds of fruit over the life of the tree. It has an efficient breeding system, each fruit producing about 800 (hermaphrodite fruit) or 1000 (female fruit) seeds, and a single tree producing hundreds of fruit in its lifetime to provide an abundance of offspring for genetic studies. A clonal propagation system is well established, which allows testing of the same genotypes in multiple environments.
The small family Caricaceae consists of six genera and 35 species, and 32 of them are dioecious. The three exceptional species are from the genera Carica and Vasconcellea, including trioecious C. papaya and V. cundinamarcensis and monoecious V. monoica. Carica papaya is the only species in the genus Carica, while Vasconcellea is the largest genus in the family including 21 species with monoecious (1), dioecious (19), and trioecious (1) all represented.
The transgenic variety SunUp female was used for the initial genome sequencing project, for its impact on the papaya industry and to avoid complications of genome assembly in the heterozygous male specific region of the Y chromosome. SunUp was developed through transformation of Sunset, which has undergone more than 25 generations of inbreeding, an ideal homozygous genotype for a genome sequencing project. The genome of SunUp female was sequenced at 3x coverage using whole genome shotgun (WGS) approach with Sanger sequencers. It was assembled into contigs containing 271 Mb and scaffolds spanning 370 Mb including embedded gaps. The estimated residual heterozygosity of SunUp is 0.06%, confirming its highly inbred nature. The papaya genome was estimated to contain 23,151 genes, about 25% less than Arabidopsis. Papaya has an AT-rich genome, with an overall G+C content of 35.3%, and consists of about 52% repetitive sequences. Papaya is the 5th angiosperm genome to be sequenced and the first transgenic crop to be characterized at the whole genome level.
We are currently pursuing funding for finishing the papaya genome and re-sequencing selected genomes from all sex genera in Central America, South America, and Africa.

Genomic basis of biomass yield in sugarcane
As a C4 plant, sugarcane has been recognized as one of the world’s most efficient crops in converting solar energy into chemical energy. Sugars can be easily converted to ethanol and carbon dioxide by bacteria or yeast through fermentation. The current momentum on biofuel and energy crop research coupled with the rapid advancement on genomic technology make sequencing the sugarcane genome a real possibility. In collaboration with colleagues in the Energy Bioscience Institute and the Hawaii Agriculture Research Center, we will study the genetic base of biomass production using interspecific F2 populations of sugarcane. We will also study genome wide gene expression patterns among energy cane (biomass production), sugar cane (sugar production), and their wild relatives S. robustum and S. spontaneum.

Figure 3. High biomass yield clone 09-9180 (in the middle) from an interspecific F2 population after 6 months growth in Kunia, Oahu, Hawaii.
Flower development in spinach (Spinacia oleracia)
In spinach, sex typically segregates equally between male and female plants he sexually dimorphic character is controlled by a locus on the largest chromosome. The locus controls sex in an XY fashion, meaning that XX is female and XY are male. Interestingly, the sex chromosomes are not always sufficient to enforce perfect male or female sex types in spinach plants. Monoecy is commonly observed in many accessions. We are studying the X/Y inheritance system with respect to monoecious genes. Also, in addition to sex determination, we are studying flowering time and other diverse floral development traits found in the spinach germplasm.
Genome evolution in Caffea
Coffee is the world’s second most widely traded commodity after oil. We are studying the forces driving genome evolution in the high quality tetraploid Arabica coffee (Caffea arabica) in comparison with the genomes of its progenitor diploid species, C. canephora and C. eugenioides. We will sequence selected bacteria artificial chromosomes (BACs) of diploid C.canephora and two sets of ortholog BACs of tetraploid C. arabica sub-genomes. Genome wide sequence analysis will be carried out in collaboration with colleagues at the International Coffee Genome Sequencing Consortium and the International Coffee Genome Network.

Figure 4. Parental Arabica coffee varieties of an F2 mapping population. From top left clockwise: Tall Mokka - High cupping quality, small leaves and beans; Catimor - Low cupping quality, large leaves and beans; Comparison of leaf size; Comparison of bean size.
NSF Project:
GEPR: The origin and evolution of sex chromosomes in Caricaceae
PI: Ray Ming, University of Illinois at Urbana-Champaign
co-PIs: Paul Moore, Hawaii Agriculture Research Center
Qingyi Yu, Texas A&M University
Richard Moore, Miami University
Collaborator: Francis Zee, USDA-ARS, Pacific Basin Agricultural Research Center
Intellectual merit and summary: Sex expression in papaya (Carica papaya L.; Family Caricaceae) is controlled by loci in the male specific region of the Y chromosome (MSY). Unlike other ancient sex chromosomes, the MSY of papaya is about 2-3 million years old and restricted to a small region about 8 Mb. Thus, the papaya sex chromosomes offer the opportunity to understand the early events in sex chromosome evolution. Moreover, sex reversal mutants indicate that the two sex determination genes are confined to a small region of about 1 Mb that is being sequenced, which makes the identification of sex determination genes feasible. Our long-term objective is to understand the evolutionary mechanisms governing the formation and divergence of sex chromosomes. Our specific objectives are: 1) Identify the sex determination genes in C. papaya. We will focus on the candidate genes in the 1 Mb deleted region of the male to female sex reversal mutants to identify the two sex determination genes; 2) Elucidate the origin of sex chromosomes in Caricaceae. We will perform a comparative genomic analysis in six species from three genera with diverse breeding systems and determine whether the sex chromosomes of papaya share a similar evolutionary history with the homologous regions in these sister species. 3) Determine the forces that have shaped genomic evolution of papaya sex chromosomes. We will distinguish between competing evolutionary hypotheses concerning the deterioration of the MSY by conducting population genomics analyses across the entire region and the corresponding region on the X chromosome. 4) Engineer true breeding hermaphrodite papaya varieties. This will improve papaya productivity while lowering cultivation costs. We will transform the sex determination gene promoting stamen development into female plants of leading papaya cultivars to produce hermaphrodite varieties without the Yh chromosome.
Links:



