![[titlebar]](../cyberlab.gif)
![[titlebar]](../cyberlab.gif)
What we're going to do now is give you some experimental results and let you interpret them, so let's jump right in. You have performed Restriction Digestion and Agarose Gel Electrophoresis on a plasmid you purified, using 3 different Restriction Enzymes, and the gel is shown below. Unfortunately, you forgot to label your tubes or keep good records, and the only things you can remember about the experiment are that your standards are in Lane 5 and your uncut control is in Lane 1, and that you loaded roughly the same amount of total DNA in your sample lanes (1-4). Hey, at least you remembered that much!
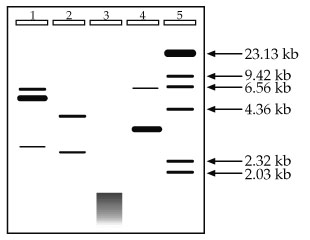
1. Given no other information and using no math, approximately how big is your original plasmid?
2. How many times did the enzyme used in Lane 2 digest the plasmid? Does the data seem reasonable? What is the likely number of base pairs this enzyme recognizes?
3. When DNA appears as a messy, continuous band as it does at the bottom of Lane 3, rather than independent, discreet bands, the effect is known as smearing. What are some likely explanations for the smearing detected in Lane 3? You should be able to come up with at least two.
4. How many times did the enzyme used in Lane 4 digest the plasmid? Does the data seem reasonable?
2. How many times did the enzyme used in Lane 2 digest the plasmid? Does the data seem reasonable? What is the likely number of base pairs this enzyme recognizes? The enzyme digests the plasmid in two places. It is important to think about the state of the DNA before digestion. The DNA used in this experiment was a plasmid, and plasmids are circular. If you cut a circle once, you get one linear fragment. You must cut it a second time to get 2 linear fragments like in Lane 2. The data does seem reasonable because if you add up the approximate sizes of the resulting fragments (roughly 4 kb and 2.5 kb), you get the original size of 6.5 kb. Lastly, it is likely that the enzyme used recognizes a sequence of 6 bases. 6-cutters, if you'll recall, cut an average of once every 4,096 bases. This is just an average, however, so in this case where we have a piece of DNA 6,500 bp long, cutting twice is very reasonable.
3. What are some likely explanations for the smearing detected in Lane 3? In general terms, smearing is when you have many bands together close enough in size that you cannot distinguish between adjacent bands (i.e., no resolution). With beginning molecular biologists, the most likely reason for the smearing is contamination by some stray nuclease that degraded the DNA into dozens, hundreds, or even thousands of little pieces. Another beginning mistake is to use the wrong buffer, wrong temperature, or wrong conditions. Any or all of these could make the enzyme behave badly, including cutting away at your DNA at multiple, random sites. However, as you do more and more experiments like this, personal error becomes less of a concern and you need to start thinking in terms of the science. If this experiment was performed without significant error, the likely explanation is that a 4-base cutter was used. Cutting an average of once every 256 bases in a 6.5 kb plasmid yields roughly 25 fragments, all smaller than the original. It is unlikely that one could see 25 individual fragments of such a small size, and the smearing pattern is probably what would be detected.
4. How many times did the enzyme used in Lane 4 digest the plasmid? Does the data seem reasonable? If you said twice, you are correct, but let's see if you were correct for the right reasons. In question 2, it was pointed out that to get two fragments from a circular piece of DNA, you need two cuts. So far so good. It was also mentioned that the total size of the resulting DNA fragments must add up to the original size. Uh oh--they don't, do they? Looking at the gel you see one band approximately 6.5 kb and one large band at roughly 3 kb. Does 6.5 + 3 = 6.5? Not in this class.
Science doesn't lie, it's just sometimes hard to interpret. So break it down. Is there anything significant about 6.5 kb? Yes, it's the size of the original plasmid. This, plus the fact that there is a band in the uncut control (Lane 1) which migrates to the same position, should suggest to you that not all of your DNA was digested (a common occurrence). This leaves the band around 3 kb. Could that band be 3.2 or 3.3 kb instead of 3.0? Easily. Unless we plot a standard curve, we're just approximating anyway. Is there anything significant about 3.3 kb? Yes, it's about half of our original sample. If the enzyme cut the plasmid into two roughly equal sized pieces, those pieces would run about the same, and would likely be indistinguishable on a gel. This is further supported by the information about this experiment which states that roughly equal amounts of DNA were loaded into Lanes 1-4. Notice how much darker the 3 kb band in Lane 4 is than the bands in Lane 2. There is twice as much DNA in that band than there is in either of the bands in Lane 2, and the data supports this conclusion.